Thread
Published!
Reproducibility Project: Cancer Biology
@eLife
* 193 experiments planned, 50 completed. Challenges in transparency, sharing, & implementation doi.org/10.7554/eLife.67995
* Replication effect sizes were 85% smaller on avg than original effects doi.org/10.7554/eLife.71601
1/
Reproducibility Project: Cancer Biology
@eLife
* 193 experiments planned, 50 completed. Challenges in transparency, sharing, & implementation doi.org/10.7554/eLife.67995
* Replication effect sizes were 85% smaller on avg than original effects doi.org/10.7554/eLife.71601
1/
This project began in 2013, had 200 contributors, was led by Tim Errington, Dir of Research @OSFramework.
Today's release includes 2 capstone papers, 2 commentaries, & the final experiments adding to the 29 Registered Reports & 17 Replication Studies.
cos.io/rpcb
Today's release includes 2 capstone papers, 2 commentaries, & the final experiments adding to the 29 Registered Reports & 17 Replication Studies.
cos.io/rpcb
1st capstone paper summarizes the challenges of conducting replications. We started with 193 experiments from 53 high-impact papers, completed 50 expts:
* data unavailable
* inadequate methods description
* reagents not shared
* variability in author help
doi.org/10.7554/eLife.67995
* data unavailable
* inadequate methods description
* reagents not shared
* variability in author help
doi.org/10.7554/eLife.67995
Figure 1 summarizes challenges in transparency, sharing, and implementation that escalated time, costs, and reduced feasibility.
Some obvious fixes: fulsome methods, open data & materials, efficient material transfer process, normalize sharing.
doi.org/10.7554/eLife.67995
Some obvious fixes: fulsome methods, open data & materials, efficient material transfer process, normalize sharing.
doi.org/10.7554/eLife.67995
We used Registered Reports (cos.io/rr/) with @elife to maximize quality of replications. This meant submitting the protocols for peer review prior to doing the research, & receiving in-principle acceptance to publish regardless of outcomes to reduce publication bias.
2nd capstone paper summarizes the 158 replication effects from the 50 completed experiments.
Effects are nested in experiments, experiments nested in papers. Report shows all levels (similar results). This thread focuses on effects, see paper for others. doi.org/10.7554/eLife.71601
Effects are nested in experiments, experiments nested in papers. Report shows all levels (similar results). This thread focuses on effects, see paper for others. doi.org/10.7554/eLife.71601
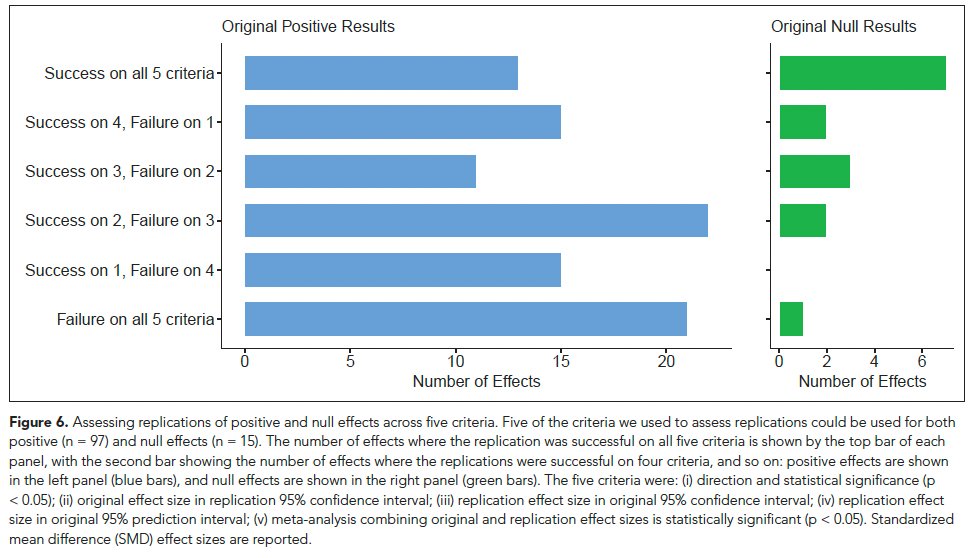
Replication success rates on 5 criteria across + and - original results:
Same direction & p<.05 (47%)
Original effect size in the rep 95% CI (25%)
Rep effect size in the orig 95% CI (48%)
Rep effect size in the 95% PI (61%)
Meta-analysis of original and replication p<.05 (63%)
Same direction & p<.05 (47%)
Original effect size in the rep 95% CI (25%)
Rep effect size in the orig 95% CI (48%)
Rep effect size in the 95% PI (61%)
Meta-analysis of original and replication p<.05 (63%)
Counting successes across 5 different criteria, 40% of original positive effects replicated successfully on a majority of them, 80% of original negative effects replicated successfully on a majority of them.
doi.org/10.7554/eLife.71601
doi.org/10.7554/eLife.71601
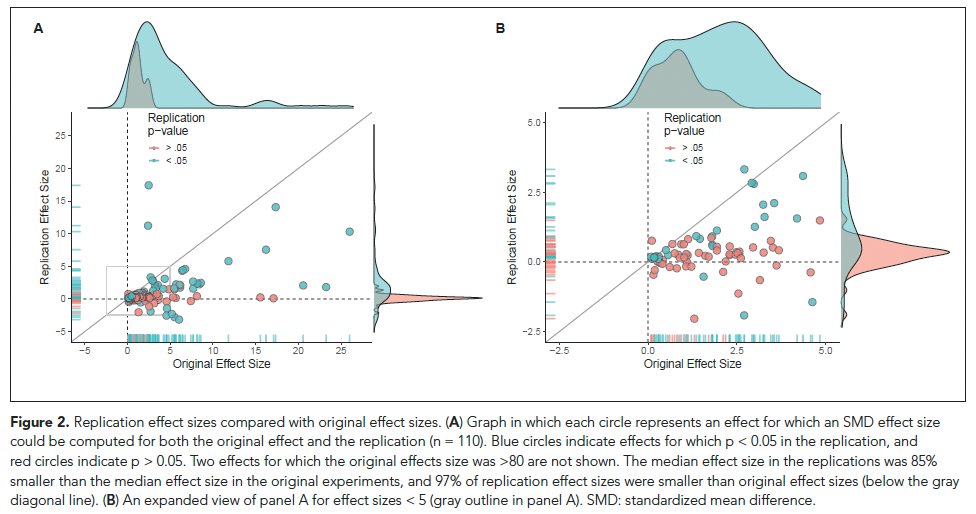
92% of the replication effect sizes were smaller than the originals. And, the replication effect sizes were 85% smaller on average than original effects.
Below diagonal replications smaller than originals. Right panel is zoom in on gray box. Blue p<.05
doi.org/10.7554/eLife.71601
Below diagonal replications smaller than originals. Right panel is zoom in on gray box. Blue p<.05
doi.org/10.7554/eLife.71601
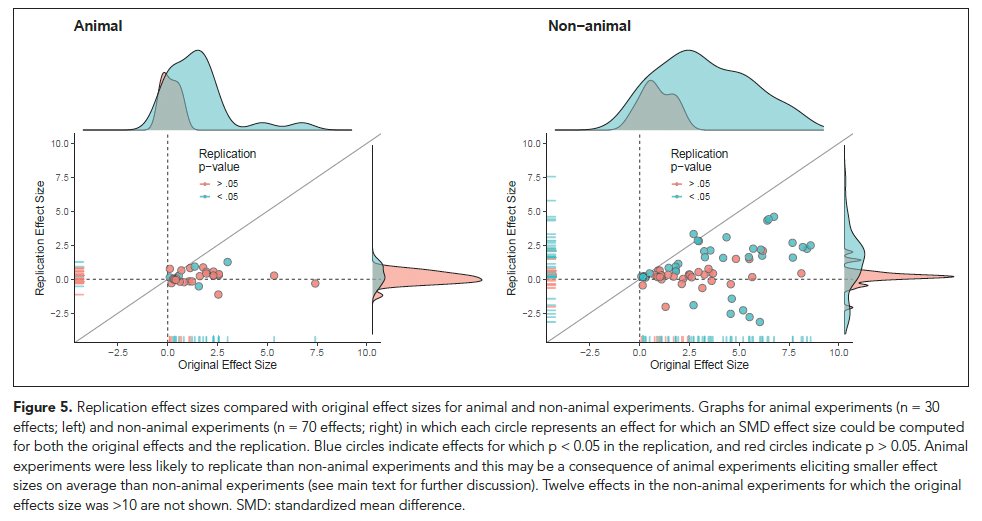
There's high interest in animal experiments given that they are often the onset to translation. Just 12% of animal replications showed p<.05 in same direction as original. But, decline in effect size was similar magnitude as non-animal (~80%). Animal ES were smaller in originals.
Acknowledgments interlude:
Tim Errington did a career's worth of effort leading this project.
@elizabethiorns and @mrgunn were the genesis of this effort.
@Arnold_Ventures, especially @JohnArnoldFndtn @LauraArnoldFdn were the supporters, early and taking a risk on us.
Tim Errington did a career's worth of effort leading this project.
@elizabethiorns and @mrgunn were the genesis of this effort.
@Arnold_Ventures, especially @JohnArnoldFndtn @LauraArnoldFdn were the supporters, early and taking a risk on us.
@stuartbuck1 was a key champion of us and of the project.
200 contributors made contributions along the way and many suppliers provided reagents to support the project. They are acknowledged here: www.cos.io/rpcb-contributors
So many thanks to so many that made this possible.
200 contributors made contributions along the way and many suppliers provided reagents to support the project. They are acknowledged here: www.cos.io/rpcb-contributors
So many thanks to so many that made this possible.
There is much more to add on the meaning and implications of these findings. For now, links:
Overview www.cos.io/rpcb
eLife collection elifesciences.org/collections/9b1e83d1/reproducibility-project-cancer-biology
data, materials, code osf.io/collections/rpcb/discover
Papers elifesciences.org/articles/67995
elifesciences.org/articles/71601
Overview www.cos.io/rpcb
eLife collection elifesciences.org/collections/9b1e83d1/reproducibility-project-cancer-biology
data, materials, code osf.io/collections/rpcb/discover
Papers elifesciences.org/articles/67995
elifesciences.org/articles/71601
Here is a companion thread unpacking the meaning and implications of these findings.